NCAR Climate System Model
Home |
Site Map |
Search |
Up One
The NCAR Ocean Model User's Guide,
Version 1.4
The NCAR Oceanography Section
1998 © UCAR/NCAR
Preface
The NCAR CSM Ocean Model (NCOM) is based on the GFDL 's Modular Ocean
Model (MOM) 1.1 global oceanic general circulation model with
substantial modifications to include improved mesoscale tracer
transport, boundary layer mixing, and surface forcing. This document
is intended to support configurations of the model which are
relatively similar to those that we use at NCAR. The details of the
model are described in greater depth within the
NCOM Technical Note.
We expect that this User's Guide will improve over time. Towards
this end your constructive comments are most welcome. Please send any
such comments to
hecht@ncar.ucar.edu.
Table of Contents
1. How to Run the Model
The NCAR CSM Ocean Model Job Scripts
Two job scripts are provided by the NCAR CSM ocean model developers:
one which compiles the ocean model and creates a binary to be used in
the coupled model system, and another which compiles and executes the
stand-alone ocean model. If the source code has been obtained from the
NCOM web page and has been unpacked, then
these two scripts will be found under the names SCRIPTS/Coupled/ocn.setup.csh
and SCRIPTS/Stand_Alone/stand-alone.csh.
Both scripts are intended to be self explanatory and are designed
to reduce the possibility of set-up errors, but because there are
certain features which may need some additional explanation, this
section provides supplementary documentation. We note that the model
has been run extensively within the NCAR environment on CRAY vector
processing machines. Although some effort has been invested towards
making the model run on other architectures, one must currently
consider the model to be unsupported on these other architectures.
Starting and Restarting the NCAR CSM Model
There are several procedures for starting and restarting the NCAR CSM
ocean model, depending on the options chosen by the user. Originally,
the CSM model had only one method for starting the model and one for
restarting it, but the subsequent addition of an automatic restart
option and an option to create a new run based on restart files from
another case increased the number of start/restart procedures.
The MOM 1.1 produced a single restart file, containing all the
necessary time levels of information. A major disadvantage of a
single restart file is that the file size gets unmanageably large as
the model resolution becomes finer. Consequently, the NCAR CSM ocean
model produces three restart files. The R2 restart file contains the
time information, barotropic streamfunction variables, and the
accumulated ice formation (when sflxiceflx is defined). The
required two time levels of the potential temperature, salinity, and
horizontal velocity fields are stored in two of the R3, R4, or R5
restart files. The empty restart file is determined automatically by
the code itself based on the number of time levels stored internally.
The autorestart feature was added for the user's convenience: it
allows the same job script to be submitted for multiple continuation
runs, completely unaltered. The user need not provide restart file
names in the script; rather, this information is read from a
"pointer" file and the ocean model, during execution, then reads the
most recent restart files from the disk location specified in the
pointer file. The autorestart option is selected by setting the
script-control variable autorestart = yes, which in turn adds
autorestart to the list of all preprocessor options.
Whether or not the autorestart feature is selected, the standard
method of starting an initial stand-alone CSM ocean model run is to
set the script variable initial_run = yes and provide initial
conditions for the starting date of the model integration, for which,
in the stand-alone model, mean January Levitus values have typically
been used. The length of the model integration is controlled by
setting numsteps, the total number of timesteps for the run.
When initial_run = yes, the job script sets the model variables
nb=0, restart-flag = false, and nlast =
numsteps.
Because there are situations in which modelers want to start a new
model run from initial conditions provided from a prior run, but want
to reset the starting time, a variant start-up option for the
stand-alone ocean is available: set initial_run = yes and
nb=0, but set restart_flag = true, and provide the names
of the R2, R3, R4, and/or R5 restart files.
The method used to continue a stand-alone model run depends on
whether or not the autorestart option is selected. If
autorestart is not in effect, the model run is continued by setting
initial_run = no, nb = itt, the timestep number from the
last restart file, restart_flag = true, autorestart = no,
and by providing the names of the restart files, i.e., the R2, R3, R4,
and/or R5 files in the appropriate part of the job script. For each
subsequent continuation run, the user must set nb = itt, and
provide the most recent restart-file names. The script will
automatically set the value of nlast to nb +
numsteps.
Continuing a stand-alone model run with the autorestart option
is simple; the user needs only to set initial_run = no
and autorestart = yes. The user may decide to reset the value of
numsteps during subsequent continuation runs, but if not, then
all continuation runs may be initiated by submitting and resubmitting
this continuation script entirely unaltered. The user should note that
the script sets nlast to numsteps in this case; however,
the autorestart preprocessor option activates code which sets
nlast to the last timestep of the previous run, itt +
nlast, thus effectively setting the total integration length to
numsteps.
Continuing a coupled ocean-model run is even simpler, because the
autorestart option is required, and the master job script (the
".nqs" script) controls whether the run is an initial,
branch or continuation run. This allows the same ocean
script to be used, unaltered, for starting and for continuing a
simulation.
Timing and Memory Requirements
Because time requirements for a model integration depend on the
particular options chosen, it is impractical to provide precise model
timing and memory information for all configurations. However, such
information from the 152x111x45 (nominally 2 degree) and 102x74x25
(nominally 3 degree) models with the options presented in this
document will provide a useful starting estimate for users who select
other model options or run on other computers.
Memory requirements for the ocean model increase with the number of
processors selected. Commonly, we run the models at NCAR on Cray C90,
YMP, or J90 computers, with either 8 or 16 processors. The 152x111x45
model requires 32Mw of memory on a 16-processor machine and 23Mw of
memory on an 8-processor machine. The figures for the 102x74x25
model are 11Mw and 8Mw, respectively. When the preprocessor option
time_average is chosen, the model memory requirements for the
152x111x45 model increase to 38Mw of memory on a 16-processor machine
and 29Mw of memory on an 8-processor machine.
NCAR CSM ocean model timing requirements and cost comparisons with
other global ocean-circulation models are discussed in detail in the
timing section at the ocean-model website
(http://www.cesm.ucar.edu/models/ocn-ncom). Timing figures are closely
linked to the options selected. For our configuration, each
152x111x45 model timestep uses approximately 4 cpu seconds on a Cray
C90, 9 cpu seconds on a Cray YM9, and 19 cpu seconds on a Cray
J90. The comparable figures for the 102x74x25 model are approximately
1, 2.4, and 5 cpu seconds, respectively.
For a detailed, up-to-date summary of changes to the ocean model,
its options and set-up scripts since our last public code release,
please refer to the section "What's New in NCOM" on the NCOM web page. We plan to periodically update
this page, so it a good idea for NCAR CSM ocean-model users to check
this page from time to time.
Time Manager
The time-manager subroutine, tmngr, has evolved at NCAR over
several years, and was eventually substantially rewritten in order to
incorporate these changes in an orderly way. In a departure from the
MOM 1.1 time manager, all time-dependent decisions in the NCAR CSM
ocean model are based on an integer timestep counter, itt, and
the corresponding elapsed model time in seconds, ttsec. Some MOM
1.1 variables which are based on the number of model days have integer
timestep-number counterparts in the CSM ocean model, but others were
eliminated during the restructuring. Many features of the MOM 1.1
time manager remain in the CSM version, although the underlying
computations have changed. Choices of a simple or Julian calendar,
allowing leap years or not, and an arbitrary year length are all
available.
New features have been added. Because model I/O has been customized
to the NCAR computing environment, new time-manager variables have
been added to control the writing and storing of files. Restart and
history files are independently controlled, and may be written at the
end of each calendar month or year. Alternatively, or additionally,
files may be written after a specified number of timesteps. Other new
time-manager variables include logical flags which signal the present
timestep is the end of a day, month, or year, and integer values of
the present second, minute, and hour of the day and day, month, and
year. All of these variables are available for use throughout the
model upon the inclusion of the files switch.h and
ctmngr2.h.
The ocean model will also write history and restart files whenever
the coupler does so. The timing of the writing of ocean history and
restart files may, if desired, be left entirely to the control of the
coupler, when the model is run in a coupled mode.
Controls for the model integration (starting and ending timesteps,
the frequency of sampling for time-averaging, the frequency of writing
diagnostics and history and restart files, and communication
information for the Flux Coupler) are all set in the namelist
contrl, which is located in the file model.data in the
stand-alone job script and ocn.parm in the coupled-model job
script. Default values for some, but not all, of these variables are
defined in subroutine blkdta.F.
Namelist contrl members include:
- nb (integer)
- the beginning timestep number. Set nb = 0 for an initial run, nb = itt for a standard continuation run, and nb > 0 for an autorstart continuation run.
- nlast (integer)
- the last timestep number of the model integration; valid only for stand-alone integrations.
- ntime_avg_freq (integer)
- the number of timesteps between the accumulation of data used in time averaging.
- write_at_reg_int (logical)
- if true, this option causes history files, restart files, and diagnostics to be written at regular intervals. Variables which must be set when this option is selected include:
- ndgnstc (integer)
- the number of timesteps between diagnostic calculations (global energetics and regional momentum and tracer term balances).
- ndmph1 (integer)
- the number of timesteps between the archiving of each model history file. Selecting ndmph1 as a multiple of nwrth1 allows for multiple writes onto one history file.
- nfacdg (integer)
- governs the frequency of printing the diagnostic computations in the subroutines diag.F and diag2.F ( i.e., nfacdg*ndgnstc timesteps).
- nprnstf (integer)
- the number of timesteps between the printing of diagnostic stream-function information ( mscan, resmax, and island values).
- nrestrt (integer)
- the number of timesteps between the creation of restart files (R~files). Restart files are sent to the NCAR mass store immediately after creation.
- ntime_avg_dump (integer)
- the number of timesteps in the time-averaging interval. This parameter also controls the writing of the time-average history files, and should be an integer multiple of ntime_avg_freq. This is only valid when the logical switch write_at_reg_int is set true.
- ntravg (integer)
- the number of timesteps between the creation of regional tracer averages.
- ntsi (integer)
- the number of timesteps between the printing of on-line integral analyses and timestep information.
- nwrth1 (integer)
- the number of timesteps between the writing of model history files.
- write_at_eom (logical)
- if true, this option causes all history files, restart files, and diagnostics to be written at the end of each month. All variables related to write_at_reg_int ( nwrth1, etc.) are ignored unless write_at_reg_int is also true.
- write_at_eoy (logical)
- if true, this option causes all history files, restart files, and diagnostics to be written at the end of each year. All variables related to write_at_reg_int ( nwrth1, etc.) are ignored unless write_at_reg_int is also true.
The ocean-model user can further control the timing of the history, restart, and diagnostics output by selecting the write_at_eom and write_at_eoy suboptions:
- write_at_eom_diags (logical)
- print diagnostics at the end of each month.
- write_at_eoy_diags (logical)
- print diagnostics at the end of each year.
- write_at_eom_history (logical)
- write history files at the end of each month.
- write_at_eoy_history (logical)
- write history files at the end of each year.
- write_at_eom_restart (logical)
- write restart files at the end of each month.
- write_at_eoy_restart (logical)
- write restart files at the end of each year.
The user can mix "eom" and "eoy" suboptions
(e.g., write_at_eom_history = .true.,
write_at_eoy_restart = .true., write_at_eom_diags =
.true.); any conflict in the options is resolved in subroutine checks
in favor of the "eoy" option.
The selection of the length of regular and leap years, the calendar
type (Julian or equally-spaced months), and specification of the
starting model time are all controlled in the namelist tminfo,
which is also set in the file model.data. At the beginning of
a model run, the starting time is specified as integer values of
day0:month0:year0 and hour0:minut0:second0. For example, a
starting date of September 3, year 5 at 3:45:15pm would be initialized
as year0=5, month0=9, day0=3, hour0=15, minut0=45, sec0=15. If
the model is being restarted from restart files, these initial time
values will be ignored, and the restart time will instead be read from
the character variable stamp, which is read from the restart
files. Variables in tminfo include:
- yrnorm (real)
- the length of a normal year, in days.
- yrleap (real)
- the length of a leap year, in days. Select yrleap = yrnorm if no leap year is desired.
- julian (logical)
- if true, selects the Julian calendar. If false, each month will contain yrnorm/12 days, and, even if specified, leap years will not be allowed.
- sec0 (integer)
- number of seconds; acceptable values: 0-59
- minut0 (integer)
- number of minutes; acceptable values: 0-59
- hour0 (integer)
- number of hours; acceptable values: 0-23
- day0 (integer)
- day number; acceptable values: 1-31
- month0 (integer)
- month number; acceptable values: 1-12
- year0 (integer)
- year number; acceptable values: 0,1,...
New options have been added to the tsteps namelist, mainly for use in the coupled-ocean model. Instead of specifying dtts in seconds, the user may instead set
- compute_dt (logical)
- if true, the ocean model
computes the timestep dtts at run-time: dtts = 1.0/
n_dt_per_day.
- n_dt_per_day (integer)
- the number of timesteps in one day.
For coupled-ocean integrations, the user may set either
- nflux (integer)
- the number of ocean-model timesteps between flux exchanges with the flux-coupler, or
- n_com_per_day (integer)
- the number of communications (flux exchanges) with the flux coupler per model day. This option is useful in conjunction with compute_dt = .true.
If both of these variables are set and one conflicts with the other, the ocean model will stop with an error message. Note that the relationship among the variables is nflux = n_dt_per_day/n_com_per_day.
Preprocessor Options
This is a list of preprocessor options in effect for standard CSM
ocean runs.
| |
152x111x45 stand-alone ocean |
152x111x45 coupled ocean/atmosphere |
102x74x25 stand-alone ocean |
102x74x25 coupled ocean/atmosphere |
| Numerical
options | congrad5pt | congrad5pt | congrad5pt | congrad5pt |
|
islands | islands | islands | islands |
|
fourfil | fourfil | fourfil | fourfil |
|
northpole | northpole | northpole | northpole |
|
taperhrzvd | taperhrzvd | taperhrzvd | taperhrzvd |
|
upwind3 | upwind3 | |
| Vertical sub-grid-scale mixing | implicitvmix | implicitvmix | implicitvmix | implicitvmix |
|
kmix | kmix | kmix | kmix |
| Horizontal and isopycnalsub-grid-scale mixing | consthmix | consthmix | consthmix | consthmix |
|
isopycmix | isopycmix | isopycmix | isopycmix |
|
isopycmixspatialvar | isopycmixspatialvar | isopycmixspatialvar | isopycmixspatialvar |
| Surface forcing options | sflxcombo | sflxpvm | sflxcombo | sflxpvm |
|
sflxcomboavan | sflxpvmicetilt | sflxcomboavan | sflxpvmicetilt |
|
sflxiceflx | sflxiceflx | sflxiceflx | sflxiceflx |
|
sflxsw | sflxsw | sflxsw | sflxsw |
|
restormed | | |
| Boundary conditions | cyclic | cyclic | cyclic | cyclic |
| Input/Output | diskless | diskless | diskless | diskless |
| History | netcdf* | netcdf* | netcdf* | netcdf* |
|
time_average | time_average | time_average | time_average |
| Model performance and monitoring | multitasking | multitasking | multitasking | multitasking |
|
readchunk | readchunk | |
|
timing | timing | timing | timing |
| Conveniences | autorestart | autorestart | autorestart | autorestart |
|
readrmsk | readrmsk | readrmsk | readrmsk |
| Hardware options | CRAY | CRAY | CRAY | CRAY |
*We plan to use this option in all CSM
production runs; it has only been used in short test cases at this time.
2. List of Preprocessor Options
Supported Options
It is our intention that all of the following options be functional,
and correct. The particular combinations of options which have been
tested with some thoroughness are those which appear in the two job
scripts, included with the code release. Users who configure the model
differently are advised to proceed with some caution. We would
appreciate being informed of any problems which may arise.
Numerical Options
The user must select the first option and any or all of the remaining
options.
- congrad5pt
- Use 5-point conjugate-gradient elliptic
solver for barotropic streamfunction.
- islands
- Include islands in relaxation topography.
- fourfil
- Fourier filter the prognostic variables in the longitudinal direction at high latitudes.
- northpole
- No island at the North Pole; see the
section Numerical and Coupling Improvements in the
NCOM Technical Note.
- taperhrzvd
- Taper horizontal and isopycnal mixing
coefficients at high latitudes; see the section Numerical and Coupling Improvements in the
NCOM Technical Note.
- upwind3
- Use third-order upwinding option; see the
section Numerical and Coupling Improvements in the
NCOM Technical Note.
Vertical Sub-grid-scale Mixing Schemes
One of the first two options must be selected. If kmix is
selected, then implicitvmix must also be selected. Multiple
sub-options, which are indented following the major option, may be
selected.
- constvmix
- Constant vertical profiles for fkpm
and fkph.
- implicitvmix
- Implicit vertical mixing; convective
adjustment is bypassed.
- kmix
- Mixing scheme based on the K-profile planetary
boundary layer scheme by Large, McWilliams, and Doney; see the
section KPP Boundary Layer Mixing Scheme in the
NCOM Technical Note.
- kmixcheckekmo
- Check Ekman Layer and Monin-Obukhov
length scales.
- kmixnori
- The vertical viscosity and diffusivity below
the boundary layer do not depend on the local Richardson number, but
are set to the background values, fkpm and fkph.
- kmixdd
- Include double-diffusion contribution to vertical
diffusivity.
Horizontal and Isopycnal Sub-grid-scale Mixing Schemes
One of the first two options must be selected. In addition, isopycmix
may be selected.
- biharmonic
- Constant coefficient biharmonic
horizontal mixing (
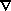 4).
4).
- consthmix
- Constant coefficient Laplacian horizontal
mixing (
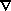 2).
2).
- isopycmix
- Mesoscale eddy parameterization; see the
section Parameterization of Mesoscale Eddies in the
NCOM Technical Note.
This only applies to the tracer equations, and it is recommended that
the coefficient ah is zero when this option is chosen.
- isopycmixspatialvar
- Taper the isopycnal mixing coefficient
to zero when isopycnal slopes become steep. This option is valid
only when isopycmix is set.
Surface Forcing Options
Only one of the first four major options should be selected.
sflxiceflx and sflxsw may be selected in addition. The
forcing of the stand-alone ocean model is documented in
Large et al. (1997), J. Phys. Oceanogr. 27, 2418-2447
(available in
pre-print form as a postscript file of 3.25MB).
- sflxcombo
- Specify type of tracer forcing data: This is
a combination of air-sea fluxes (based on air temperature, specific
humidity, solar short wave flux, cloud fraction, precipitation, and
windspeed) and restoring terms (due to restoring fields of SST and
SSS).
- sflxcomboavan
- Compute annual averages of several
surface tracer fluxes for diagnostic purposes.
- sflxpvm
- Provide surface fluxes via the Flux Coupler.
- sflxpvmicetilt
- Compute the tilt of the sea surface;
necessary for some ice models.
- restorst
- Restore sea-surface temperature and salinity back
to specified values globally. By default, mid-month values are
assumed, and linear interpolation in time is performed. A single
restoring time scale is applied at all surface points.
- sflxiceflx
- Check for ice formation in the uppermost
kmxice layers in the ocean model, change temperature and salinity
if ice forms or melts, and keep track of previously formed ice. Can
be used for ocean only model runs. Should be used for coupled runs.
- sflxsw
- Treat solar short wave flux differently from all
other surface heat fluxes: this flux appears as a right-hand-side
term in sourct and is allowed to penetrate the water with an
exponentially decreasing absorption factor. The Jerlov water type,
jwatertype, is specified in blkdta.F.
Boundary Conditions
- cyclic
- Impose cyclic longitudinal boundary conditions.
Input/Output
- crayio
- Use I/O based on Cray getwa and putwa commands for out-of-core computations.
- diskless
- Simulate disk storage using memory array for in-core computations.
History
- inst_hist
- Create instantaneous snapshots of the model
fields, forcing and poleward transports.
- netcdf
- Use netCDF file format for history files (if
not selected, then native binary representation is used).
- time_average
- Create time averages of the model
fields, forcing and poleward transports (see the section
Numerical and Coupling Improvements in the NCOM Technical
Note).
Model Performance and Monitoring
- multitasking
- Allow parallel processing via latitude bands.
- readchunk
- Specify the chunk size for each set of latitudes so that the
computational load is balanced among all of the processors.
Presently, this option is valid only when using the 152x111x45 model, with
ntasks=4,8, or 16; the chunks are defined in blkdta.F.
- keepterms
- Use arrays to retain terms in momentum and
tracer equations, instead of recomputing via statement functions.
- testcfl
- Test whether any velocity exceeds the local CFL
criterion by a factor of cflcrt.
- timing
- Compute the time spent per gridpoint per timestep;
report cpu and wall-clock times. The implementation is specific to
CRAY machines running UNICOS.
Conveniences
- autorestart
- Provide an automatic restart from the latest
model restart files.
- readrmsk
- Read in region information from the file "regionmask".
Hardware Options
- NOMSS
- Do not use the NCAR Mass Storage System subroutines
msread and mswrite to move history and restart files to archival
storage; rather, use replacement subroutines which use Unix commands
via "call system" or "ishell" to move these files to a more
permanent location. Note that the user may be required to provide a
local version of system, a subroutine which interacts with the local
operating system from the FORTRAN program, if the user's local system
does not support ishell.
- TASKCOMMON
- Replace each instance of "TASKCOMMON"
with "task common" on Cray and HP hardware; replace it with "common"
on all others.
- CRAY
- Indicates the ocean model is being run on a Cray
computer; "ishell" is used for all system calls from the model and
Cray pointers are used in the subroutine step.
- HP
- Indicates that the model is being run on a
computer which recognizes HP optimization commands.
Internal Options
- hmixalreadyset
- An internal option set in the files fdifm.h and fdift.h
- ioalreadyset
- An internal option set in the file odam.F
Passive Tracers
- ideal_age
- Include a passive age tracer which is
set to zero at the surface, and incremented at a rate of 1 unit per
year in all subsurface layers.
Unsupported Options
The following options exist in the NCAR CSM ocean model, but they are
not supported by the NCAR CSM developers and should be activated by
the user only after careful testing to ensure correctness.
- congrad9pt
- Use 9-point conjugate-gradient elliptic solver
for barotropic streamfunction.
- constvmixdmxl
- Enhance vertical diffusivity in the top 50m (3
or 4 layers).
- constvmixvmxl
- Enhance vertical viscosity in the top 50m (3 or
4 layers).
- coupled_ocean_only
- Run the equivalent of the stand-alone
ocean model in coupled mode, through the flux-coupler. This option
requires the existence of special climatological ice, land, and
atmospheric component models, which are presently unavailable.
- coupled_ocean_ice
- Run the ocean model coupled with an
active ice model through the flux-coupler. This option requires
special climatological land and atmospheric component models, which
are presently unavailable.
- extras
- Provide extra diagnostic information.
- fio
- Fortran direct-access I/O.
- firfil
- Finite-impulse filter.
- insitu
- Compute polynomial expansion coefficients for the
equation of state using in-situ temperature.
- knudsen
- Use Knudsen equation of state for sea water.
- multitaskqueue
- Monitor the status of the NCAR Cray
multitasking queue; shut down and exit upon receiving the proper signal.
- nlhmix
- Nonlinear horizontal sub-grid-scale mixing.
- nohilats
- Drop metric nonlinear terms in the momentum equation.
- ppvmix
- Use Pacanowski and Philander vertical mixing.
- sflxdataflx
- Specify all fluxes from data:
forcing data consist of non-solar heat flux, solar heat flux, and net
freshwater flux.
- sflxtharm
- Use annual and first two harmonics of the
seasonal cycle for restoring of the surface tracer fields.
- sflxwharm
- Use annual and first two harmonics of the
seasonal cycle for wind stress forcing.
- skipland
- Perform computations only over ocean points.
- symmetry
- Symmetric boundary condition about the northern model boundary.
- tcvmix
- Mellor-Yamada turbulence closure scheme.
- lalg
- tcvmix suboption.
- leq
- tcvmix suboption.
Incomplete Options
The following options which appear in the NCAR CSM ocean model are in
fact incomplete within the publicly released code, as they are still
under development and evaluation.
- bbl_adv
- Use the advective bottom boundary layer
parameterization of Beckmann and Döscher.
- bbl_diff
- Use the diffusive bottom boundary layer
parameterization of Beckmann and Döscher.
- carbon14
- Single-tracer carbon 14.
- co2
- Solubility-driven carbon dioxide.
- runoff
- Receive river runoff from the flux coupler
and distribute it into the ocean model.
- sigmac
- Invoke the sigma-coordinate model.
- upperocean
- Invoke the sigma-coordinate upper-ocean
model.
3. Parameter Values from Published Simulations
Here we list some of the most important model parameters and the
settings of those parameters, as the model was configure for several
published works. The 152x111x45 cases are documented in several of the
papers which appear in the May issue of the Journal of Climate
(Vol. 11, No. 6). The 102x74x25 case is documented in
Large et al. (1997), J. Phys. Oceanogr. 27, 2418-2447
(available as in
pre-print form as a postscript file of 3.25MB).
The default parameter values can be found in the Fortran block data
files blkdata.F and kmixbdta.F. Within the job shell script a file
model.data is created which contains a subset of the model parameters
and these values override the default values in the block data files.
Many of these can be changed at run time by specifying their values in
the namelists which are defined in the file nldefs.h.
We note that the parameter values listed below are historical. The
currently recommended values are those appearing in the most recent
public release of NCOM, available at the NCOM
web page.
Stand-alone 152x111x45 model configuration (accelerated run)
| Variable | Value |
|---|
| Horizontal viscosity | am = 8.0 e+8 |
| Background vertical viscosity | fkpm = 10.0 |
| Horizontal diffusivity | ah = 0.0 |
| Background vertical diffusivity | fkph = 0.3 |
| Bottom friction | cdbot = 0.001 |
| Eddy-induced advection coefficient | athkdf = 0.6 e+7 |
| Isopycnal diffusivity | ahisop = 0.6 e+7 |
| Inverse maximum slope | slmxr = 100.0 |
| KPP critical bulk Richardson number | Ricr = 0.3 |
| KPP critical local Richardson number | Riinfty = 0.7 |
| KPP shear vertical viscosity | vvcric = 50.0 |
| KPP shear vertical diffusivity | vdcric = 50.0 |
| KPP convective vertical viscosity | vvclim = 1000.0 |
| KPP convective vertical diffusivity | vdclim = 1000.0 |
| Strong heat restoring under ice | restQp = 386.0 |
| Strong salt restoring under ice | restFp = 2.77 |
| Weak ocean heat restoring | restQm = 0.0 |
| Weak ocean salt restoring | restFm = 0.092 |
| Number of layers which can form ice | kmxice = 1 |
| Tracer timestep | dtts = 24000.0 |
| Baroclinic velocity timestep | dtuv = 2400.0 |
| Barotropic velocity timestep | dtsf = 2400.0 |
| Tracer vertical acceleration | dtxcel =
25*1.0, 4*5.0, 2*6.0,2*7.0, |
| | 2*8.0,2*9.0, 8*10.0 |
| Maximum number of scans | mxscan = 1000 |
| Barotropic convergence criterion | crit = 2.0 e+7 |
| Fraction of implicit Coriolis force | acor = 0.0 |
Coupled 152x111x45 model configuration (synchronous run)
The only changes from the 152x111x45 stand-alone parameter values are
| Variable | Value |
|---|
| Tracer timestep | dtts = 2400.0 |
| Tracer vertical acceleration | dtxcel = 45*1.0 |
and the four surface forcing restoring parameter values are
irrelevant.
Stand-alone 102x74x25 model configuration (accelerated run)
| Variable | Value |
|---|
| Horizontal viscosity | am = 3.0 e+9 |
| Background vertical viscosity | fkpm = 16.7 |
| Horizontal diffusivity | ah = 0.0 |
| Background vertical diffusivity | fkph = 0.5 |
| Bottom friction | cdbot = 0.001 |
| Eddy-induced advection coefficient | athkdf = 0.8 e+7 |
| Isopycnal diffusivity | ahisop = 0.8 e+7 |
| Inverse maximum slope | slmxr = 100.0 |
| KPP critical bulk Richardson number | Ricr = 0.3 |
| KPP critical local Richardson number | Riinfty = 0.7 |
| KPP shear vertical viscosity | vvcric = 50.0 |
| KPP shear vertical diffusivity | vdcric = 50.0 |
| KPP convective vertical viscosity | vvclim = 1000.0 |
| KPP convective vertical diffusivity | vdclim = 1000.0 |
| Strong heat restoring under ice | restQp = 386.0 |
| Strong salt restoring under ice | restFp = 2.77 |
| Weak ocean heat restoring | restQm = 0.0 |
| Weak ocean salt restoring | restFm = 0.023 |
| Number of layers which can form ice | kmxice = 1 |
| Tracer timestep | dtts = 35040.0 |
| Baroclinic velocity timestep | dtuv = 3504.0 |
| Barotropic velocity timestep | dtsf = 3504.0 |
| Tracer vertical acceleration | dtxcel = 14*1.0, 5*5.0,
6*10.0 |
| Maximum number of scans | mxscan = 1000 |
| Barotropic convergence criterion | crit = 1.0 e+6 |
| Fraction of implicit Coriolis force | acor = 0.0 |
Last modified by Matthew Hecht,
Thursday, 10-Jun-2010 14:00:51 MDT
hecht@ncar.ucar.edu
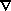 4).
4).
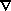 2).
2).